Welcome to the Weatheritt lab website
Our lab utilizes computational and molecular biology techniques to investigate how post-transcriptional processes, particularly alternative splicing, regulate protein function in Motor Neuron Diseases (MND) and other neurological disorders. Our goal is to uncover the root causes of these debilitating conditions and develop novel therapeutic interventions.
News
Research
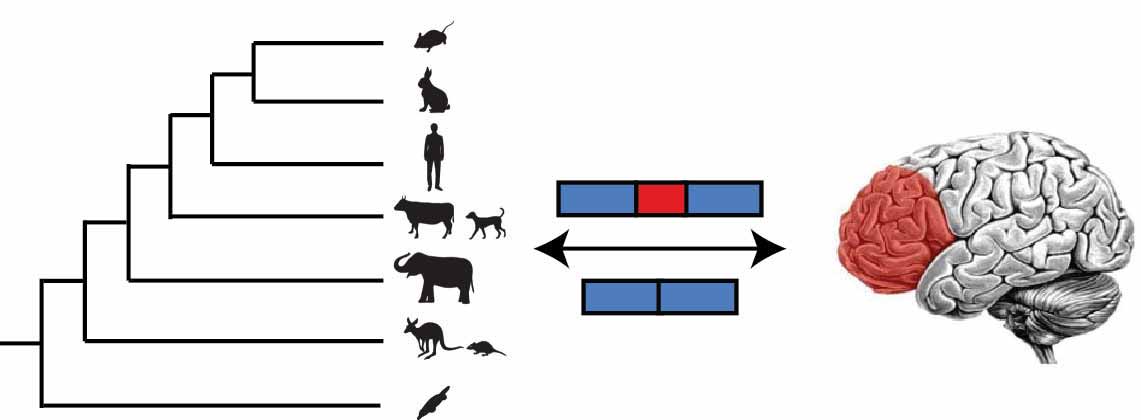
Human have approximately 86 trillion neurons. C. elegans has exactly 302 neurons. Humans and C. elegans have approximately the same number of genes. How is this complexity encoded? We are interested in investigating how
Relevant Publications:
Irimia et al. Cell 2014
Santos-Rodriguez et al. eLife 2021
Bartonicek et al. Nature 2022
What is the impact of membraneless organelles on cellular phenotypes? Does splicing regulate the function of membraneless organelles?
Relevant Publications:
Avgan et al. Genome Biology. 2019
Gueroussov et al. Cell. 2017:
Weatheritt and Babu. Science 2013:
the proteome
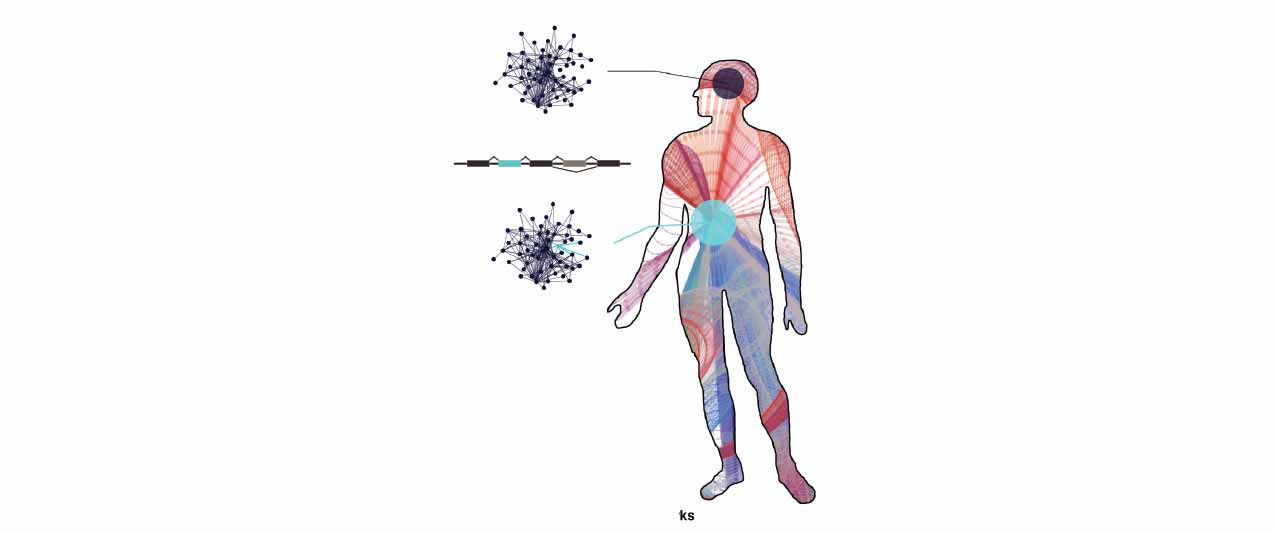
We previously demonstrated that tissue-specific alternative splicing rewires proteomic networks by targeting linear motifs within intrinsically disorder regions. How dynamic is this process? Is it regulated by cell signalling?
Relevant Publications:
Kjer Hansen and Weatheritt. NSMB. 2023
Weatheritt et al. NSMB. 2016
Sinitcyn et al. Nature Biotech
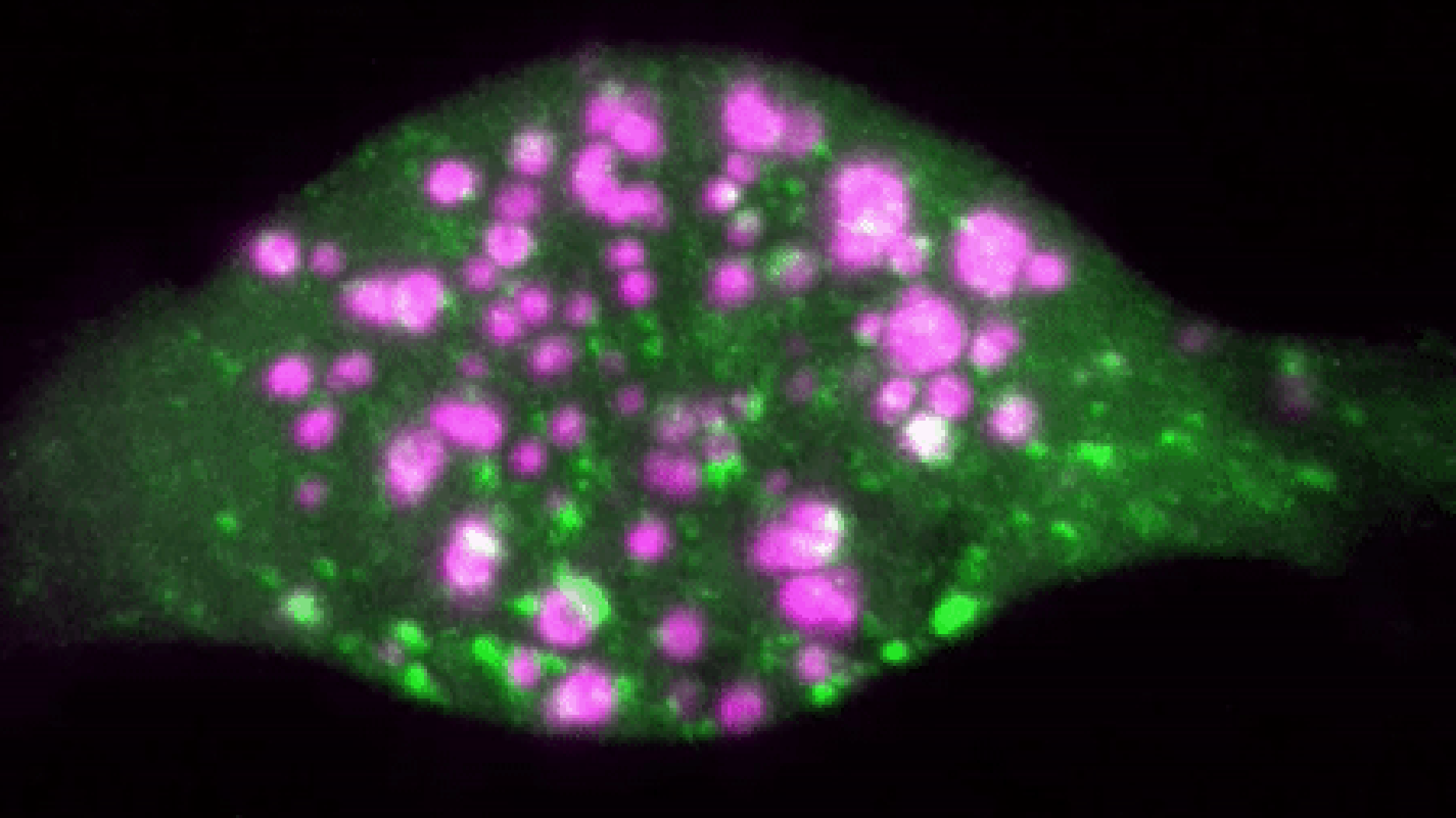
We use high content imaging studies combined with CRISPR technology to understand functional differences between proteins produced by alternative splicing.
Irimia et al. Cell 2014
Kjer Hansen and Weatheritt. NSMB. 2023
Recent Publications
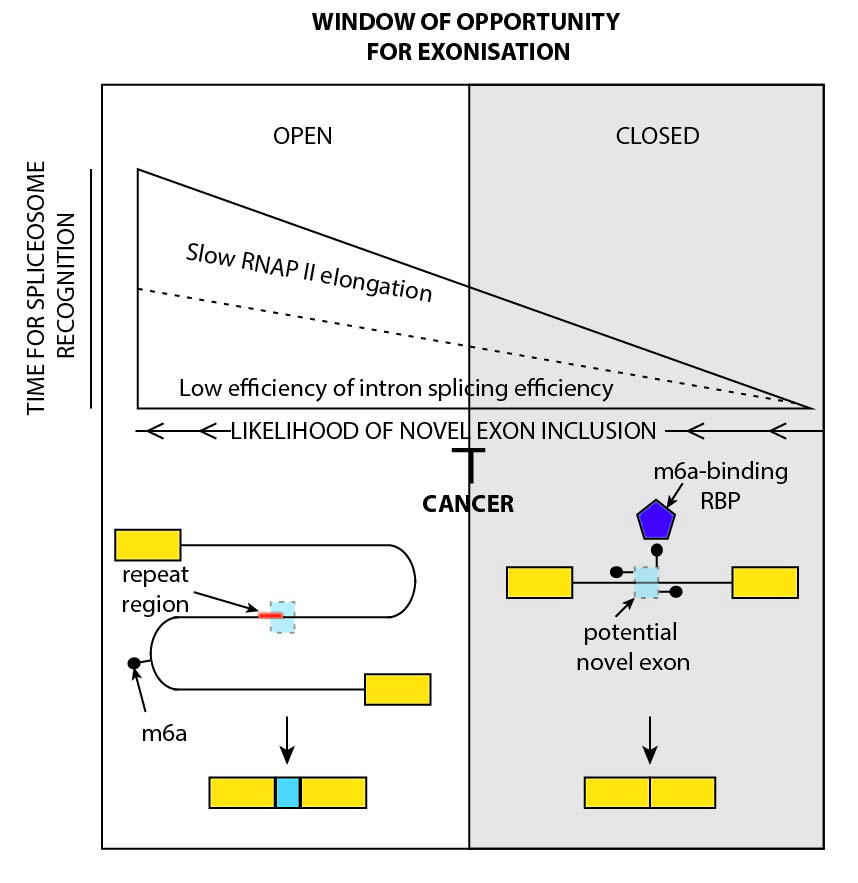
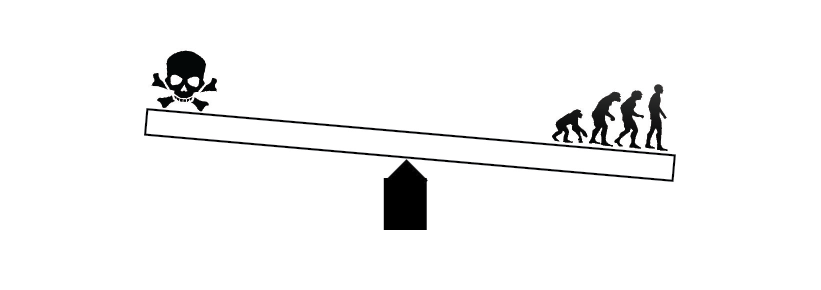
We identified a exonisation is controlled by a multilayered mechanism that modulates the "window of opportunity" for cryptic exon recognition.
Avgan et al.
Genome Biology (2019)
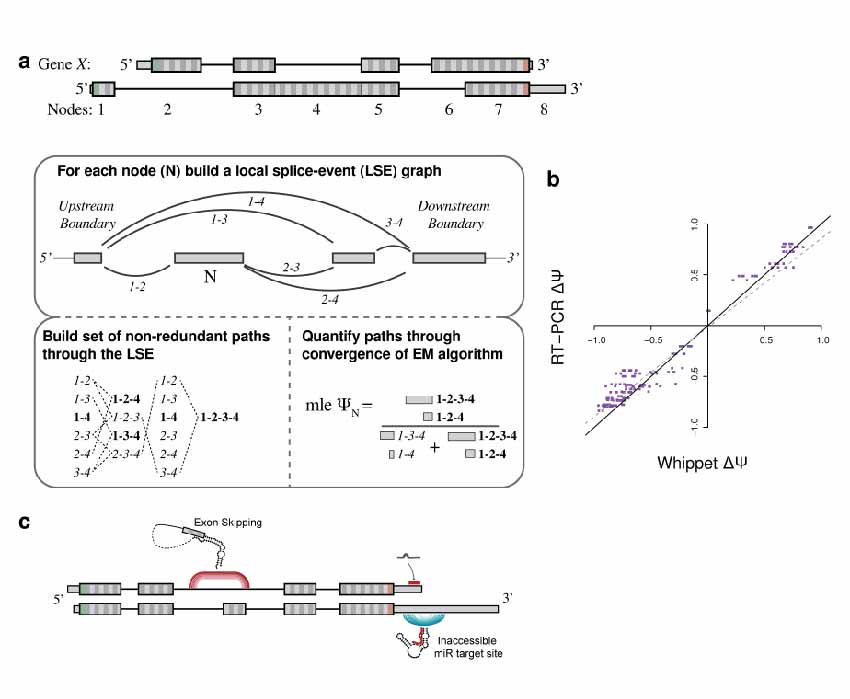
We've created a light-weight, lightening fast method for the quantitative profiling of AS from raw RNA-seq reads at the event-level (Whippet.jl). We identify >30% of human genome under complex splicing events and cancer genomes show more highly complex transcriptomes.
Sterne-Weiler, Weatheritt et al.
Molecular Cell (2018)
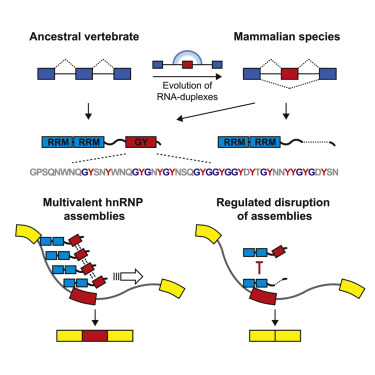
We identified a series of mammalian-specific alternative splicing events that regulate higher order protein assemblies. This mechanism expands the regulatory complexity of mammalian transcriptomes and proteomes.
Gueroussov, Weatheritt et al.
Cell (2017)
Full List of Publications
• By Journal : Cell (2), Nature (2), Science (1), Chemical Reviews (2), Nat Struc Biol (3), Mol Cell (6), PNAS (1), E-life (1), Nature Communication (1), Science Signal (1), TIBS (1), Protein Sci (1), Mol BioSyst (2), Curr Opin Stru (1), NAR (5), Bioinfomatics (1), Front Neuro (1), Hum Brain Mapp (1), Nature Review Genetics (1), Genome Biology (1), Nature Biotechnology (1), Nature Review Drug Discovery (1)
• By year : 2024 (4), 2023 (4), 2022 (5), 2021 (2), 2020 (4), 2019 (2), 2018 (1), 2017 (2), 2016 (3), 2015 (1), 2014 (7), 2013 (4), 2012 (5), < 2012 (3)
- Alternative splicing: regulation, biological significance and evolution
1) Kjer-Hansen P, Phan TG, Weatheritt RJ . Protein isoform-centric therapeutics: expanding targets and increasing specificity. Nat Rev Drug Discov. 2024 Oct;23(10):759-779. doi: 10.1038/s41573-024-01025-z
2) Kjer-Hansen P, Weatheritt RJ. The function of alternative splicing in the proteome: rewiring protein interactomes to put old functions into new contexts. Nature Structural & Molecular Biology (NSMB) 2023 Nov 30. doi: 10.1038/s41594-023-01155-9
3) Sinitcyn P, Richards AL, Weatheritt RJ , Brademan DR, Marx H, Meyer JG, Shishkvoa E, Hebert AS, Westphall MS, Blencowe BJ, Cox J, Coon JJ. Deep Human Proteome Sequencing for Global Detection of Mutations and Alternative Splicing. Nature Biotechnology 2023 Mar 23. doi: 10.1038/s41587-023-01714-x.
4) Skvortsova K, Bertrand S, Voronov D, Ducket PE, Ross SE, Magri MS, Maeso I, Weatheritt RJ , Skarmeta JLG, Arnone MI, Escriva H, Bogdanovic O. Active DNA demethylation of developmental cis-regulatory regions predates vertebrate origins Sci Adv. 2022 Dec 2;8(48):eabn2258
5) Bartonicek N, Rouet R, Warren J, Loetsch C, Santos-Rodriguez G, Walters S, Lin F, Zahra D,Blackburn J, Hammond GM, Reis ALM, Deveson IR, Zammit N, Zeraati M, Grey S, Christ D, Mattick JS, Chtanova T, Brink R, Dinger ME, Weatheritt RJ , Sprent J, King C. The retroelement Lx9 puts a brake on the immune response to virus infection. Nature 2022 Aug;608(7924):757-765.
6) Han H, Best AJ, Braunschweig U, Mikolajewicz N, Li JD, Roth J, Chowdhury F, Mantica F, Nabeel-Shah S, Parada G, Brown KR, O'Hanlon D, Wei J, Yao Y, Zid AA, Comsa LC, Jen M, Wang J, Datti A, Gonatopoulos-Pournatzis T, Weatheritt RJ , Greenblatt JF, Wrana JL, Irimia M, Gingras AC, Moffat J, Blencowe BJ. Systematic exploration of dynamic splicing networks reveals conserved multistage regulators of neurogenesis. Molecular Cell 2022 Aug 18;82(16):2982-2999.e14
7) Wang J , Weatheritt RJ , Voineagu I Alu-minating the Mechanisms Underlying Primate Cortex Evolution. Biol Psychiatry 2022 May 19:S0006-3223(22)01273-2
8) Barutcu AR, Wu M, Braunschweig U, Dyakov BJA, Luo Z, Turner KM, Durbic T, Lin ZY, Weatheritt RJ , Maass PG, Gingras AC, Blencowe BJ. Systematic mapping of nuclear domain-associated transcripts reveals speckles and lamina as hubs of functionally distinct retained introns. Molecular Cell . 2022 Feb 9:S1097-2765(21)01072-8
9) Santos-Rodriguez G , Voineagu I, Weatheritt RJ . Evolutionary dynamics of circular RNAs in primates. eLIFE 2021 Sep 20;10:e69148
10) Ha KCH, Sterne-Weiler T, Morris Q, Weatheritt RJ , Blencowe BJ. Differential contribution of transcriptomic regulatory layers in the definition of neuronal identity. Nature Communications 2021 Jan 12;12(1):335.
11) Farini D, Cesari E, Weatheritt RJ , La Sala G, Naro C, Pagliarini V, Bonvissuto D, Medici V, Guerra M, Di Pietro C, Rizzo FR, Musella A, Carola V, Centonze D, Blencowe BJ, Marazziti D, Sette C. A Dynamic Splicing Program Ensures Proper Synaptic Connections in the Developing Cerebellum. Cell Reports 2020 Jun 2;31(9):107703
12) Luck K, Kim DK, Lambourne L, Spirohn K,..., Weatheritt RJ and HURI Consortium. Nature . 2020 Apr;580(7803):402-408A reference map of the human binary protein interactome.
13) Rodrigues DC, Mufteev M, Weatheritt RJ , Djuric U, Ha KCH, Ross PJ, Wei W, Piekna A, Sartori MA, Byres L, Mok RSF, Zaslavsky K, Pasceri P, Diamandis P, Morris Q, Blencowe BJ, Ellis J. Shifts in Ribosome Engagement Impact Key Gene Sets in Neurodevelopment and Ubiquitination in Rett Syndrome. Cell Reports . 2020 Mar 24;30(12):4179-4196
14) Gonatopoulos-Pournatzis T, Niibori R, Salter EW, Weatheritt RJ , Tsang B, Farhangmehr S, Liang X, Braunschweig U, Roth J, Zhang S, Henderson T, Sharma E, Quesnel-Vallières M, Permanyer J, Maier S, Georgiou J, Irimia M, Sonenberg N, Forman-Kay JD, Gingras AC, Collingridge GL, Woodin MA, Cordes SP, Blencowe BJ. Autism-Misregulated eIF4G Microexons Control Synaptic Translation and Higher Order Cognitive Functions. Molecular Cell . 2020 Jan 25. pii: S1097-2765(20)30006-X.
15) Avgan N , Wang JI , Fernandez-Chamorro J , Weatheritt RJ . Multilayered control of exon acquisition permits the emergence of novel forms of regulatory control. Genome Biology . 2019 Jul 17;20(1):141
16) Quesnel-Vallières M, Weatheritt RJ , Cordes SP, Blencowe BJ. Autism spectrum disorder: insights into convergent mechanisms from transcriptomics
Nat Rev Genetics . 2019 Jan;20(1):51-63
- on cover!
17) Sterne-Weiler T*, Weatheritt RJ* , Best A, Ha KCH, Blencowe BJ. Whippet: an ultra fast and accurate
alignment and quantification of RNA-seq data for analysis of alternative splicing. Molecular Cell 2018 Oct 4;72(1):187-200.e6
18) Gueroussov S*, Weatheritt RJ* , Sterne-Weiler T, O’Hanlon D, Lin ZY, Gingras AC, Blencowe BJ§.
Mammalian-specific regulation of higher-order hnRNP protein assemblies controls alternative splicing. Cell
2017 Jul 13;170(2):324-339.e23.
- research highlighted by Nature Review Molecular Cell Biology
19) Han H, Braunschweig U, Gonatopoulos-Pournatzis T, Weatheritt RJ , Hirsch CL, Ha KCH,
Radovani E, Nabell-Shah S, Sterne-Weiler T, Wang J, O’Hanlon D, Pan Q, Ray D, Vizeacoumar F, Datti A,
Magomedova L, Cummins CL, Hughes TR, Greenblatt JF, Wrana JL, Moffat J, Blencowe BJ. Multilayered
control of alternative splicing regulatory networks by transcription factors. Molecular Cell . 2017 Feb
2;65(3):539-553.e7.
20) Weatheritt RJ , Sterne-Weiler T, Blencowe BJ. (2016) The ribosome-engaged landscape of alternative
splicing. Nat Struct Mol Biol. 2016 Dec;23(12):1117-1123
21) Dominguez D, Tsai YH, Weatheritt R , Wang Y, Blencowe BJ, Wang Z. (2016) An extensive program of
periodic alternative splicing linked to cell cycle progression. E-life. 2016 Mar 25;5
22) Irimia M, Weatheritt RJ , Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T,Babor M, Quesnel-Vallières
M, Tapial J, Raj B, O'Hanlon D, Barrios-Rodiles M, Sternberg MJ, Cordes SP, Roth FP, Wrana JL, Geschwind DH, Blencowe BJ. (2014) A highly conserved program of neuronal microexons is misregulatedin autistic brains. Cell . 159(7):1511-23
- perspective in Cell by Yang and Chen, and in EMBO Journal by Darnell. Research highlighted by Nature Review Neuroscience and Faculty of 1000
23) Reyes A, Anders S, Weatheritt RJ , Gibson T.J, Steinmetz L, Huber W. (2013) Drift and conservation of
differential exon usage across tissues in primate species. PNAS. 110(38)
24) Weatheritt RJ , Davey NE, Gibson TJ. (2012) Linear motifs confer functional diversity onto splice
variants. Nucleic Acids Res 40, 7123-7131
- recommended reading by Nature Review Genetics
25) Weatheritt RJ and Gibson,TJ. (2012) Linear Motifs: Lost in (Pre)Translation Trends Biochem Sci i 37,
333-341 [Review article]
- Genomic and post-transcriptional regulation
26) Latysheva NS, Oates ME, Maddox L, Flock T, Gough J, Buljan M, Weatheritt RJ , Babu MM. (2016)
Molecular Principles of Gene Fusion Mediated Rewiring of Protein Interaction Networks in Cancer.
Molecular Cell. 2016 Aug 18;63(4):579-92
27) Weatheritt RJ , Gibson TJ, Babu MM. (2014) Asymmetric mRNA localization contributes to fidelity
and sensitivity of spatially localized systems. Nat Struct Mol Biol. 21(9); 833-9
- research highlighted by Faculty of 1000 and by Nature.com
28) Weatheritt RJ , Babu MM. (2013) Evolution. The hidden codes that shape protein evolution. Science
342(6164)
- Intrinsically disordered regions and short linear motifs (SLiMs)
29) Latysheva NS, Flock T, Weatheritt RJ , Chavali S, Babu MM. (2015) How do disordered regions achieve
comparable functions to structured domains? Protein Sci. 2015 Jun;24(6):909-22 [Review article]
30) Uyar B, Weatheritt RJ , Dinkel H, Davey NE, Gibson TJ. (2014) Proteome-wide analysis of human disease
mutations in short linear motifs: neglected players in cancer? Mol BioSyst ; 10(10): 2626-42
31) Van Roey K, Uyar, Weatheritt RJ , Dinkel H, Seiler M, Budd A, Gibson TJ, Davey NE. (2014) Short
linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation.
Chemical Reviews. 114(13):6733-78 [Review article]
32) Van der Lee R, Buljan M*, Lang B*, Weatheritt RJ* , Daughdrill GW, Dunker AK, Fuxreiter M, Gough J,
Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE,
Babu MM. (2014) Classification of Intrinsically Disordered Regions and Proteins. Chemical Reviews 114
(13): 6589-631 [Review article]
33) Flock T*, Weatheritt RJ* , Latsheva NS, Babu MM§. (2014) Controlling entropy to tune the functions of
intrinsically disordered regions. Curr Opin Struct Biol. 2014 Jun;26C:62-7 [Review article]
34) Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ , Born D, Speck T, Krüger D, Grebnev G,
Kuban M, Strumillo M, Uyar B, Budd A, Altenberg B, Seiler M, Chemes LB, Glavina J, Sánchez IE, Diella
F, Gibson TJ. (2014) The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res
D259-66
35) Van Roey K, Dinkel H*, Weatheritt, RJ* , Gibson TJ, Davey NE. (2013) The switches.ELM resource: a
compendium of conditional regulatory interfaces within the intrinsically disordered proteome. Science
Signaling. 6(269): rs7
36) Weatheritt RJ , Jehl P, Dinkel H, Gibson TJ. (2012) iELM – a web server to explore short linear motif
mediated interactions Nucleic Acids Res. doi: 40, W364-9
37) Weatheritt RJ , Luck K, Petsalaki E, Davey NE, Gibson TJ. (2012) The identification of short linear motif
mediated interfaces within the human interactome Bioinformatics 28(7):976-982
38) Davey NE, Van Roey K*, Weatheritt RJ* , Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H,
Gibson TJ. (2012) Attributes of short linear motifs. Mol Biosyst 8, 268-281.
39) Dinkel H, Michael S, Weatheritt RJ , Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M,
Budd A, Jodicke L, Dammert MA, Schroeter C, Hammer M, Schmidt T, Jehl P, McGuigan C, Dymecka M,
Chica C, Luck K, Via A, Chatr-Aryamontri A, Haslam N, Grebnev G, Edwards RJ, Steinmetz MO,
Meiselbach H, Diella F, Gibson TJ. (2012) ELM--the database of eukaryotic linear motifs. Nucleic Acids
Res 40, D242-D251.
40) Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne
JC, Chica C, Seiler M, Davey NE, Haslam N, Weatheritt RJ , Budd A, Hughes T, Pas J, Rychlewski L,
Trave G, Aasland R, Helmer-Citterich M, Linding R, Gibson TJ. (2010) ELM: the status of the 2010
eukaryotic linear motif resource. Nucleic Acids Res 38, D167-80.
- Neuroscience
41) Yanashima R, Kitagawa N, Matsubara Y, Weatheritt R , Oka K, Kikuchi S, Tomita M, Ishizaki S. (2009)
Network Features and Pathway Analyses of a Signal Transduction Cascade. Front Neuroinform 3, 13.
42) Krumbholz K, Nobis EA, Weatheritt RJ , Fink GR. (2009) Executive control of spatial attention shifts in
the auditory compared to the visual modality. Hum Brain Mapp 30, 1457-146
People

EMBL Australia Group Leader
Education: University of York (MRes), EMBL Heidelberg (PhD), MRC Laboratory of Molecular Biology and Donnelly Centre (PostDoc)
Hometown: London, England
Other activities: Diving, Sailing, Squash

Lab Manager
Education: Federal University of Ouro Preto (MSc), Federal University of Minas Gerais (PhD)
Hometown: Belo Horizonte/Minas Gerais - Brazil.
Other activities: Rock Climbing, Capoeira, Yoga, dancing Forró

Research Intern
Education: University of Cambridge (MPhil) University of Huddersfield (BSc Honours)
Hometown: Leeds, England
Other activities: Exploring new places, Hiking and Reading

PhD Student
Education: King's College London (BSc & MRes)
Hometown: Windsor, UK.
Other activities: Basketball, Yoga, Attempting Ottolenghi recipes

Postdoc
Education: University of Copenhagen (BSc and MSc)
Hometown: Copenhagen, Denmark.
Other activities: Exploring newer music history, reading actual books, and coffee

Postdoctoral Fellow
Education: University of Tasmania (PhD)
Hometown: Hobart. Tasmania / Puerto Montt, Chile
Other activities: Hiking, dancing, walking my cat

Postdoctoral Fellow
Education: University of Queensland (PhD)
Hometown: India
Other activities: Hiking, dancing
Join us

Postdocs
If you are interested in our research and you have a recent PhD degree in molecular biology, proteomics/genomics, computation biology, neuroscience or bioinformatics, please send an email to Robert explaining your research interests (also include your CV and contact details of three references). We are always interested in motivated candidates. Potential candidates who are interested in joining us should be competitive to apply for external fellowships and sources of funding such as the ARC fellowship, NHMRC fellowship, Marie-Curie Fellowship or HFSP Fellowship.
Graduate students
If you are interested in joining the lab or doing a master's project, please send Robert an email (r.weatheritt@garvan.org.au) or come by his office. Also, have a look at the Garvan institute graduate programme or EMBL Australia Partnership PhD Program for information about the application process.
Undergraduate students
We may have openings for undergraduate students. If you are interested, please send Robert an email with a short explanation of your interests. Please attach your transcript and CV in the email. Also, have a look at the Garvan institute undergraduate programme for more information.
Contact
Or visit us at the Garvan:




